alessandro.ratto{a}uclouvain.be
https://twitter.com/lilissandro
Alessandro RATTO
BIO
I was born and raised in Turin, Italy, where I got a Bachelor’s degree in Chemistry and a Master’s degree in Industrial Biotechnology, two disciplines that mutually benefit each other when mixed. During an undergraduate Erasmus exchange at Universidad de La Laguna, Spain, I fell in love with Biochemistry and Protein Engineering, a passion that would later ensue in a thesis on the molecular mechanism of the first oxygen-resistant [FeFe]-hydrogenase ever discovered. That research experience, combined with an innate curiosity and an international mindset, would ultimately lead me to Belgium, where I’m now pursuing a PhD in Molecular Biology & Genetic Engineering.
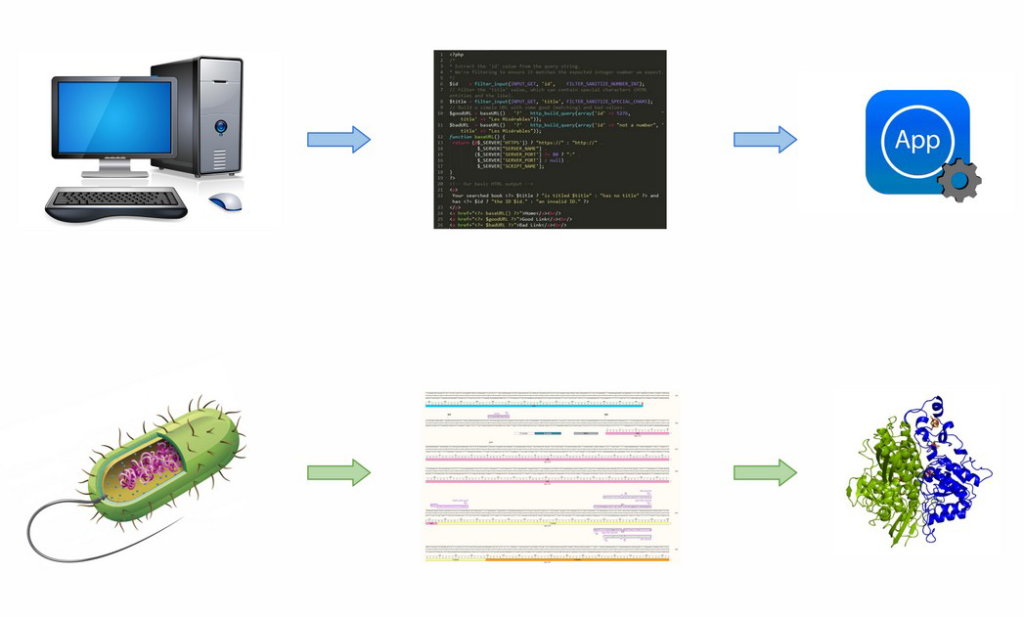
PROJECT
My PhD project falls under the EU Marie Curie Innovative Training Network (ITN) “EVOdrops”, an international, cross-institute effort focused on pushing the envelope of directed evolution + droplet-based microfluidics. These and other recent Nobel prize-winning technologies are progressively enabling scientists to treat DNA as a programming language – much like Python or C++ – to code for specific functions, in a way similar to that of digital apps. Unfortunately, the biotechnological heaven ushered in by a “programmable biocatalysis” and synthetic biology is still far away, whereas the methodologies available today are still imperfect and labour-intensive. Hence, my research focuses on making the enzyme directed evolution workflow easier, especially when it is carried out in droplet-based microfluidic setups which have the highest throughput. In particular, I design genetic “software” and “install” it in Gram+/- bacteria via genome editing techniques (such as CRISPR) in order to create host strains that externalize enzymes of interest and can therefore be used as “lysis-free” cellular platforms for protein expression and screening.