Preferences of DNA binding proteins for DNA Shape Beyond Sequence Motifs
– Jason Baby Chirakadavil
A graphical summary of the key points discussed in [1]
Brief:
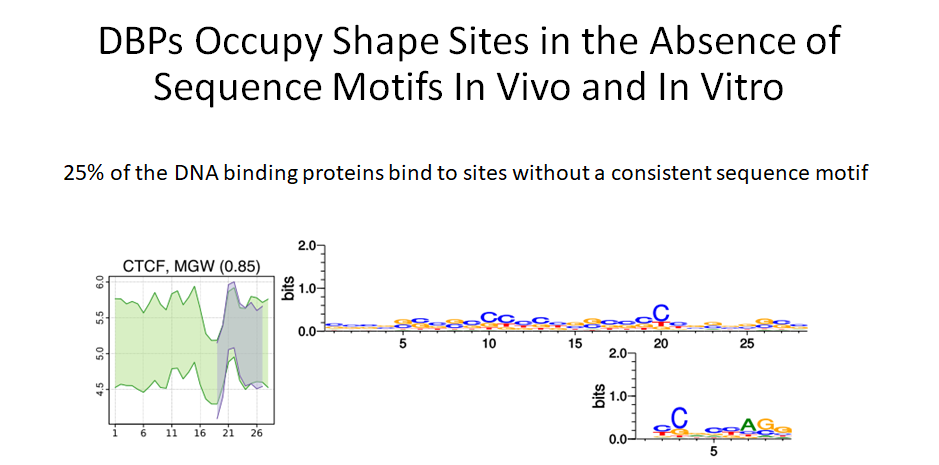
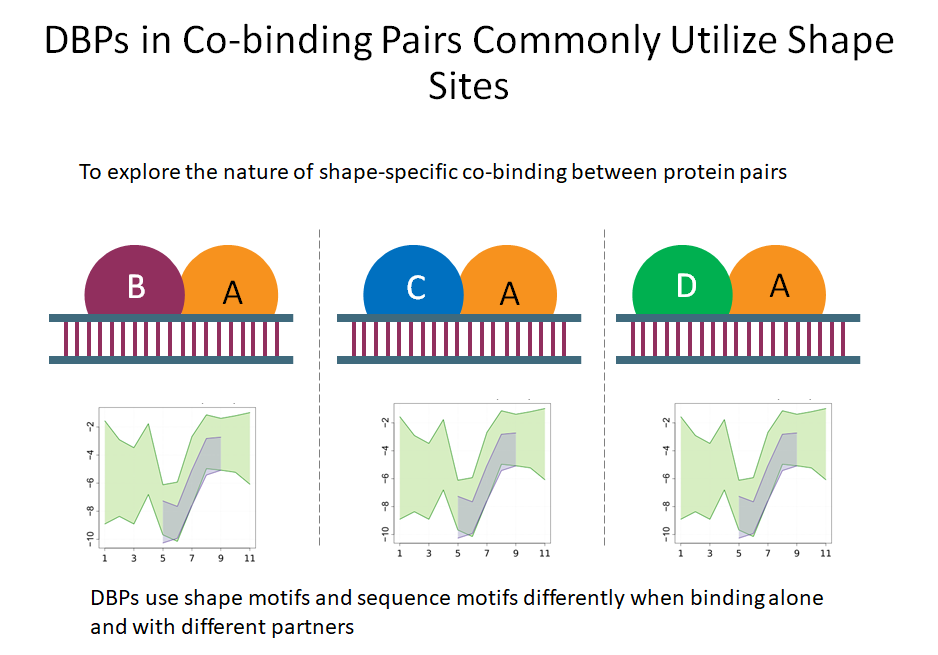
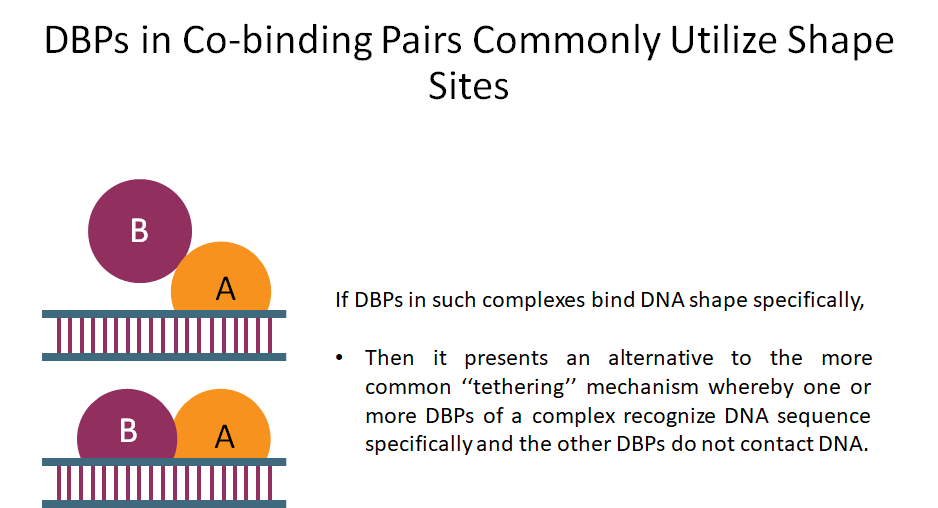
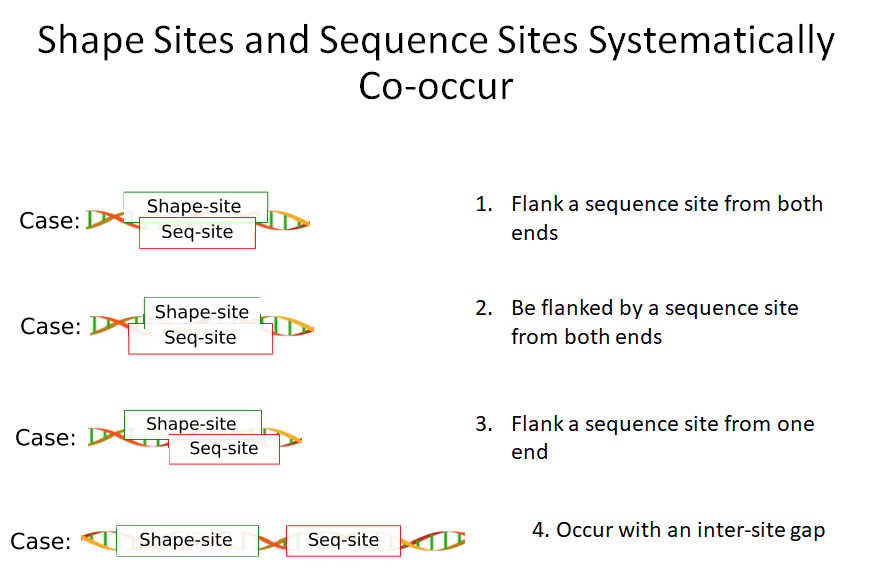
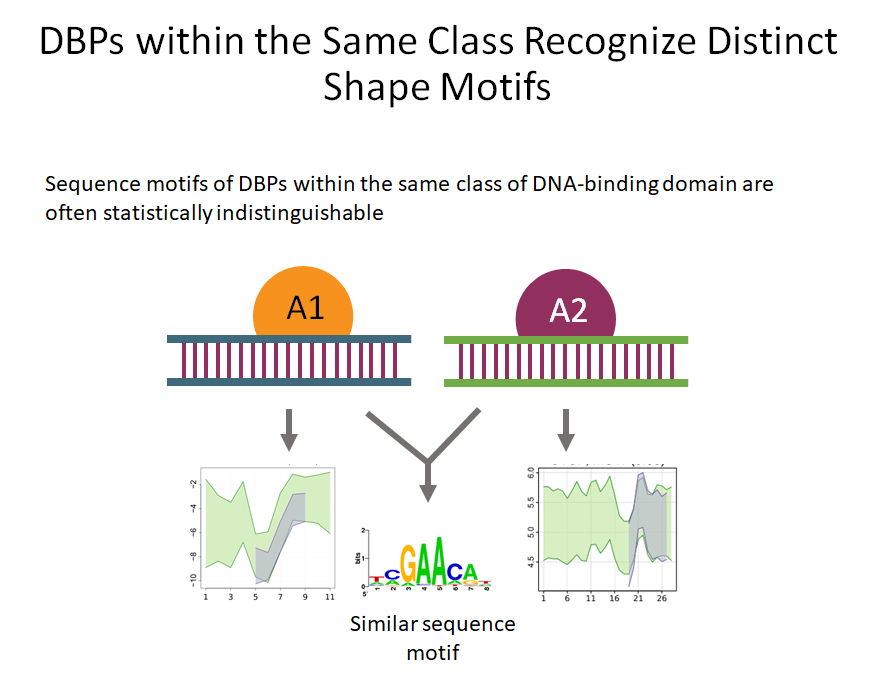
Building on the recent line of research that DNA shape encodes specificity signals for transcription factor (TF)-DNA binding, Samee et al. developed an algorithm to identify enriched patterns of DNA shape directly from DNA shape profiles of TF-bound regions. These patterns, or ‘‘shape motifs,’’ correspond well with conventional sequence motifs, but often include additional nucleotides flanking the sequence motifs. They also show that apparent mismatches to the sequence motif can have similar shape to the shape motif and are potential targets for TF binding. While the observations still need experimental validation, the work shows that motif models that capture additional structural and biophysical aspects are necessary to understand the mechanism of TF-DNA binding.
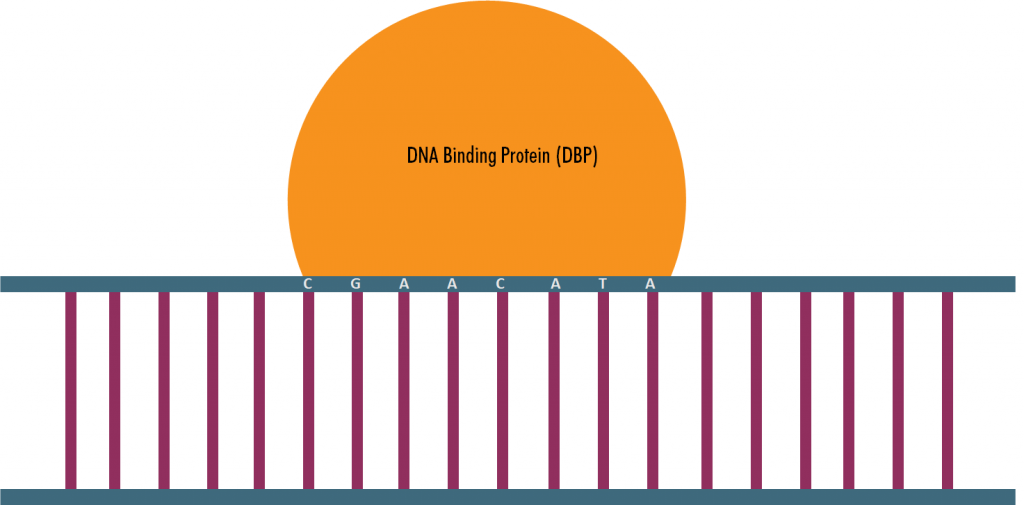
Classically we only focus on the sequence of nucleotides in DNA when looking at DNA binding proteins. But recently the specific structure of DNA sequences have been under the focus of study. Binding sequences of DNA may have specific patterns of DNA shape features which may also play a role in binding affinities.

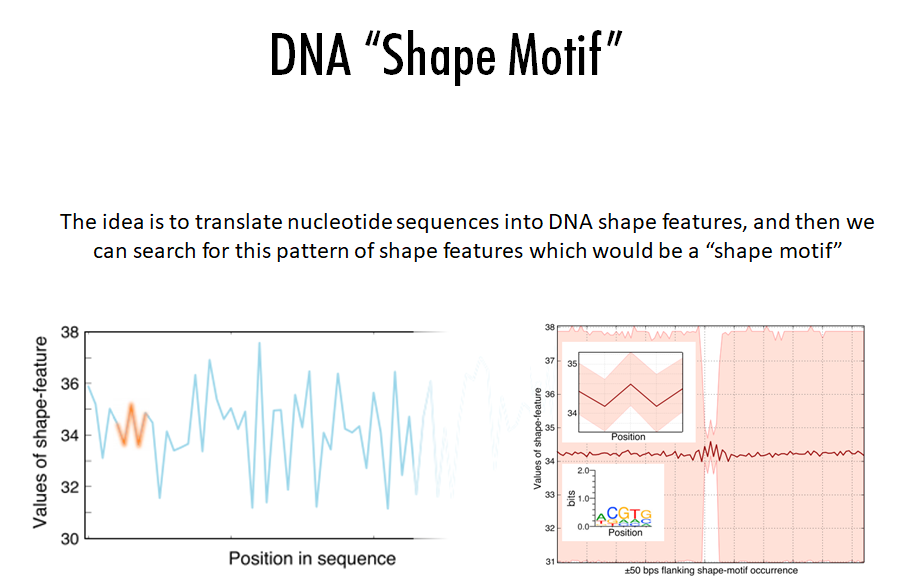
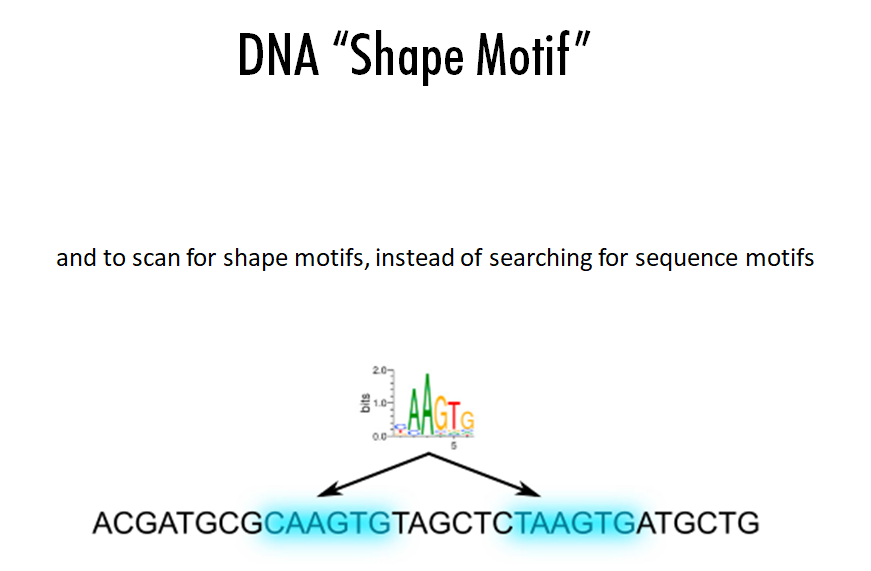

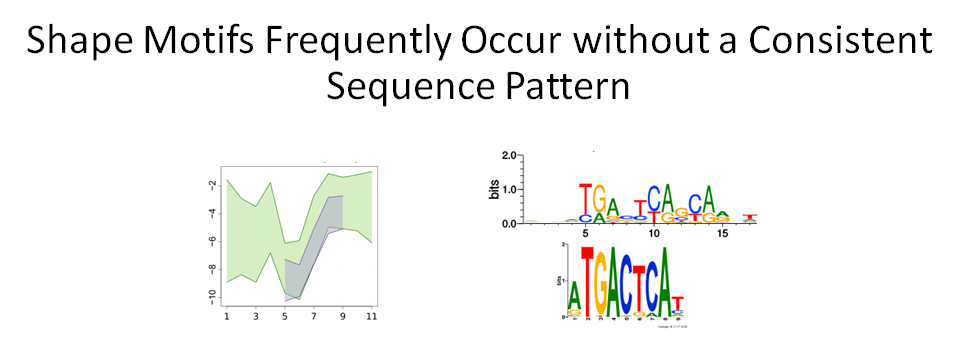
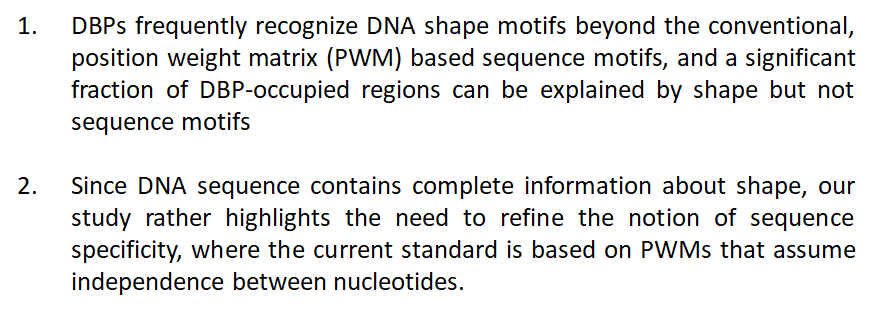
Conclusions
References:
1. Bruneau, B. G., Pollard, K. S., Samee, A. H., Bruneau, B. G., & Pollard, K. S. (2019). Article A De Novo Shape Motif Discovery Algorithm Reveals Preferences of Transcription Factors for DNA Shape Article A De Novo Shape Motif Discovery Algorithm Reveals Preferences of Transcription Factors for DNA Shape Beyond Sequence Motifs. Cell Systems, 1–16. https://doi.org/10.1016/j.cels.2018.12.001
