Laboratory of Neural Differentiation (NEDI)
Last update :
August 2024
The Onecut (OC) transcription factors
The family of Onecut transcription factors comprises three members in mammals, namely Hepatocyte Nuclear Factor 6 (HNF-6) also called OC-1, OC-2 and OC-3. These transcriptional activators contain a bipartite DNA-binding domain constituted by a single CUT domain and a homeodomain. Their high sequence similarities and their overlapping expression patterns suggest that they exert redundant functions during development and for homeostasis of adult tissues.
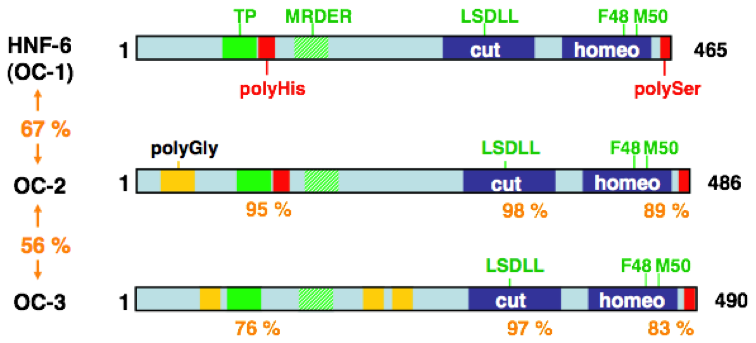
The family of Onecut transcription factors : schematic representation of the three OC proteins with the two DNA-binding domains (blue), and regions and residues that stimulate (green) or inhibit (red) their transcriptional activity. The percentage of identity of the primary sequences is indicated.
Onecut publications
Audouard E., Schakman O., René F., Huettl R.-E., Huber A.B., Loeffler J.-P., Gailly P. and Clotman F. (2012). The Onecut transcription factor HNF-6 regulates in motor neurons the formation of the neuromuscular junctions. PLoS One, 7 (12):e50509.
Audouard E., Schakman O., Ginion A., Bertrand L., Gailly P. and Clotman F. (2013). The Onecut transcription factor HNF-6 contributes to proper reorganization of Purkinje cells during postnatal cerebellum development. Molecular and Cellular Neuroscience, 56 pp. 159-168.
Chakrabarty K., Von Oerthel L., Hellemons A., Clotman F., Espana A., Groot Koerkamp M., Holstege F.C.P., Pasterkamp R.J. and Smidt M.P. (2012). Genome wide expression profiling of the mesodiencephalic region identifies novel factors involved in early and late dopaminergic development. Biology Open, doi:10.1242/bio.20121230.
Clotman, F., Jacquemin, P., Plumb-Rudewiez, N., Pierreux, C.E., Van der Smissen, P., Dietz, H.C., Courtoy, P.J., Rousseau, G.G., and Lemaigre, F.P. (2005). Control of liver cell fate decision by a gradient of TGF beta signaling modulated by Onecut transcription factors. Genes Dev 19, pp. 1849-1854.
Clotman, F., Lannoy, V.J., Reber, M., Cereghini, S., Cassiman, D., Jacquemin, P., Roskams, T., Rousseau, G.G., and Lemaigre, F.P. (2002). The onecut transcription factor HNF6 is required for normal development of the biliary tract. Development 129 pp. 1819-1828.
Espana, A. and Clotman, F. (2012). The Onecut transcription factors are required for the second phase of development of the A13 dopaminergic nucleus in the mouse. The Journal of Comparative Neurology 520 (7) pp.1424-1441.
Espana A. and Clotman F. (2012). Onecut transcription factors control development of the Locus Coeruleus and of the Mesencephalic Trigeminal Nucleus. Molecular and Cellular Neuroscience 50 (1) pp. 93-102.
Francius, C., and Clotman, F. (2010). Dynamic expression of the Onecut transcription factors HNF-6, OC-2 and OC-3 during spinal motor neuron development. Neuroscience 165 pp. 116-129.
Francius C., Clotman F. (2014). Generating spinal motor neuron diversity: a long quest for neuronal identity. Cellular and Molecular Life Science 71 pp. 813-829.
Francius C., Harris A.A., Hendricks T.J., Stam F., Barber M., Kurek D., Grosveld F.G., Pierani A., Goulding M. and Clotman F. (2013). Identification of multiple subsets of ventral interneurons and differential distribution along the rostrocaudal axis of the developing spinal cord. PLoS One, 8 (8): e70325 .
Harris A., Masgutova M., Collin A., Toch M., Hidalgo-Figueroa M., Jacob B., Corcoran L.M., Francius C. and Clotman F. (2019). Onecut factors and Pou2f2 regulate the distribution of V2 interneurons in the mouse developing spinal cord. Frontiers in Cellular Neuroscience, 13: 184.
Hodge, L.K., Klassen, M.P., Han, B.X., Yiu, G., Hurrell, J., Howell, A., Rousseau, G., Lemaigre, F., Tessier-Lavigne, M., and Wang, F. (2007). Retrograde BMP signaling regulates trigeminal sensory neuron identities and the formation of precise face maps. Neuron 55 pp. 572-586.
Jacquemin, P., Pierreux, C.E., Fierens, S., van Eyll, J.M., Lemaigre, F.P., and Rousseau, G.G. (2003b). Cloning and embryonic expression pattern of the mouse Onecut transcription factor OC-2. Gene Expr Patterns 3 pp. 639-644.
Kabayiza K., Masgutova G., Harris A., Rucchin V., Jacob B. and Clotman F. (2017)
The Onecut transcription factors regulate differentiation and distribution of dorsal interneurons during spinal cord development
Frontiers in Molecular Neuroscience, 10:157.
Landry, C., Clotman, F., Hioki, T., Oda, H., Picard, J.J., Lemaigre, F.P., and Rousseau, G.G. (1997). HNF-6 is expressed in endoderm derivatives and nervous system of the mouse embryo and participates to the cross-regulatory network of liver-enriched transcription factors. Dev Biol 192 pp. 247-257.
Lemaigre, F.P., Durviaux, S.M., Truong, O., Lannoy, V.J., Hsuan, J.J., and Rousseau, G.G. (1996). Hepatocyte nuclear factor 6, a transcription factor that contains a novel type of homeodomain and a single cut domain. Proc Natl Acad Sci U S A 93 pp. 9460-9464.
Roy A., Francius C. (equal contribution), Rousso D.L., Seuntjes E., Debruyn J., Luxenhofer G., Huber A.B., Huylebroeck D., Novitch B.G. and Clotman F. (2012). Onecut transcription factors act upstream of Isl1 to regulate spinal motoneuron diversification. Development, 139 (17) pp. 3109-19.
Samadani, U., and Costa, R.H. (1996). The transcriptional activator hepatocyte nuclear factor 6 regulates liver gene expression. Mol Cell Biol 16 pp. 6273-6284.
Stam F., Hendricks T.J. (equal contribution), Geiman E., Zhang, J., Francius C., Labosky, P., Clotman F. and Goulding M. (2012). Control of Renshaw cell inhibitory interneuron specialization by a temporally restricted transcription factor program. Development 139 (1) pp. 179-190.
Toch M. and Clotman F. (2019). CBP and p300 coactivators contribute to the maintenance of Isl1 expression by the Onecut transcription factors in embryonic spinal motor neurons. Molecular and Cellular Neuroscience, 101:103411.
Toch M., Harris A., Schakman O., Kondratskaya E., Boulland J.L., Dauguet N., Debrulle S., Baudouin C., Hidalgo-Figueroa M., Gow A., Glover J.C., Tissir F. and Clotman F. (2020). Onecut-dependent Nkx6.2 transcription factor expression is required for proper formation and activity of spinal locomotor circuits. Scientific Reports, 10(1):996.
Vanhorenbeeck, V., Jacquemin, P., Lemaigre, F.P., and Rousseau, G.G. (2002). OC-3, a novel mammalian member of the ONECUT class of transcription factors. Biochem Biophys Res Commun 292 pp. 848-854.
The OC factors are expressed in parts of the digestive tract and in the nervous system during embryonic development and in the adult. Analyses of single or compound knockout mice for the OC factors unveiled critical roles of these proteins for pancreas and liver differentiation and morphogenesis.
In the nervous system, OC proteins are detected in the spinal cord and in several regions of the encephalon excluding the cortex. In the spinal cord, they are present in subsets of motor neurons and of ventral or dorsal interneuron populations (Francius & Clotman, 2010; Francius et al., 2013; Kabayiza et al., 2017; Harris et al., 2019). Projects in our laboratory demonstrate that OC factors are required for normal development of motor neurons (Roy et al., 2012; Francius & Clotman, 2014; Toch et al., 2019) and of ventral (Stam et al., 2012; Francius et al., 2013; Harris et al., 2019; Toch et al., 2020) or dorsal (Kabayiza et al., 2017) interneurons of the spinal cord.