Background
The field of human genetics is being revolutionized by exome and genome sequencing. A massive amount of data is being produced at ever-increasing rates. Targeted exome sequencing can be completed in a few days using NGS, allowing for new variant discovery in a matter of weeks. The technology generates considerable numbers of false positives, and the differentiation of sequencing errors from true mutations is not a straightforward task. Moreover, the identification of changes-of-interest from amongst tens of thousands of variants requires annotation drawn from various sources, as well as advanced filtering capabilities.
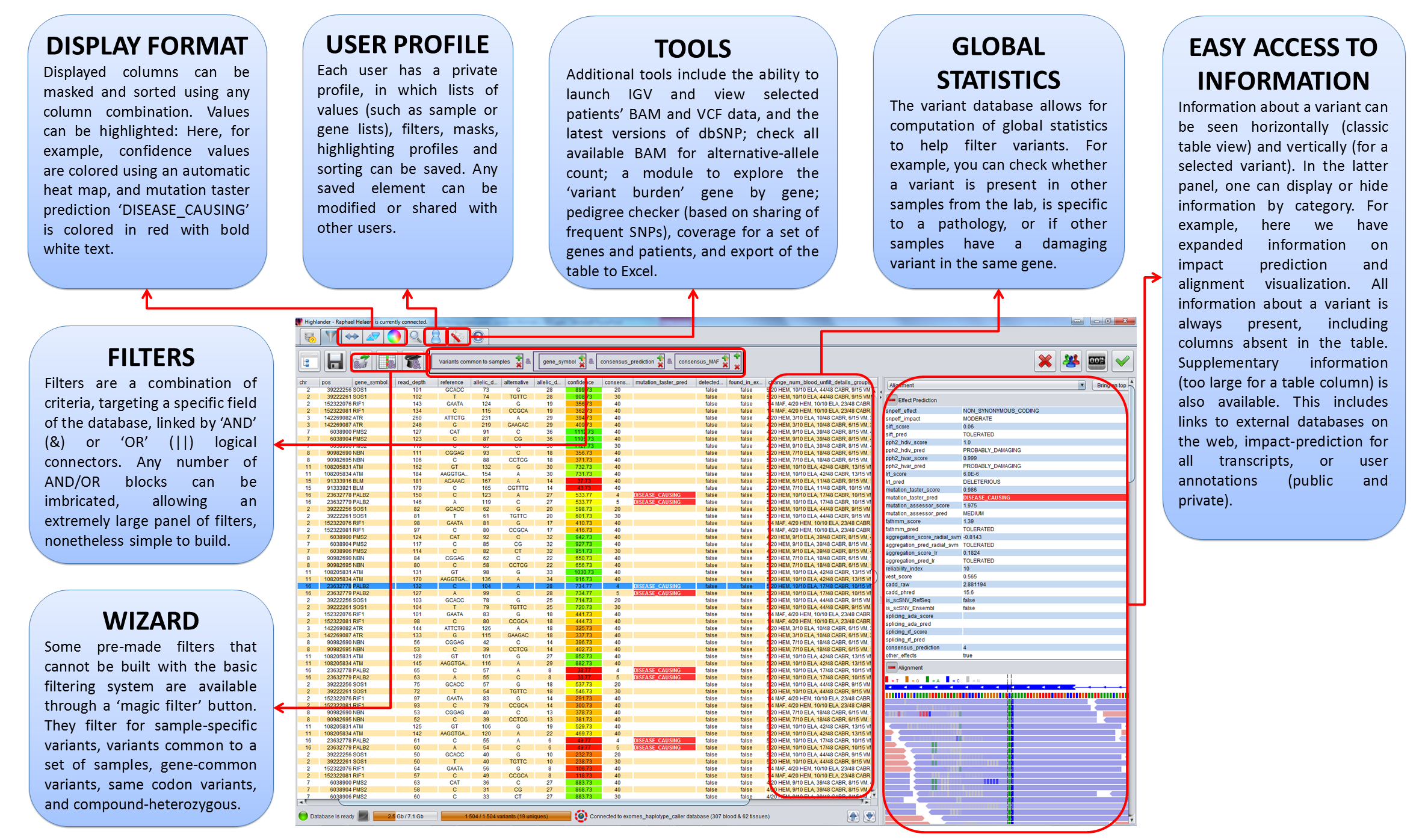
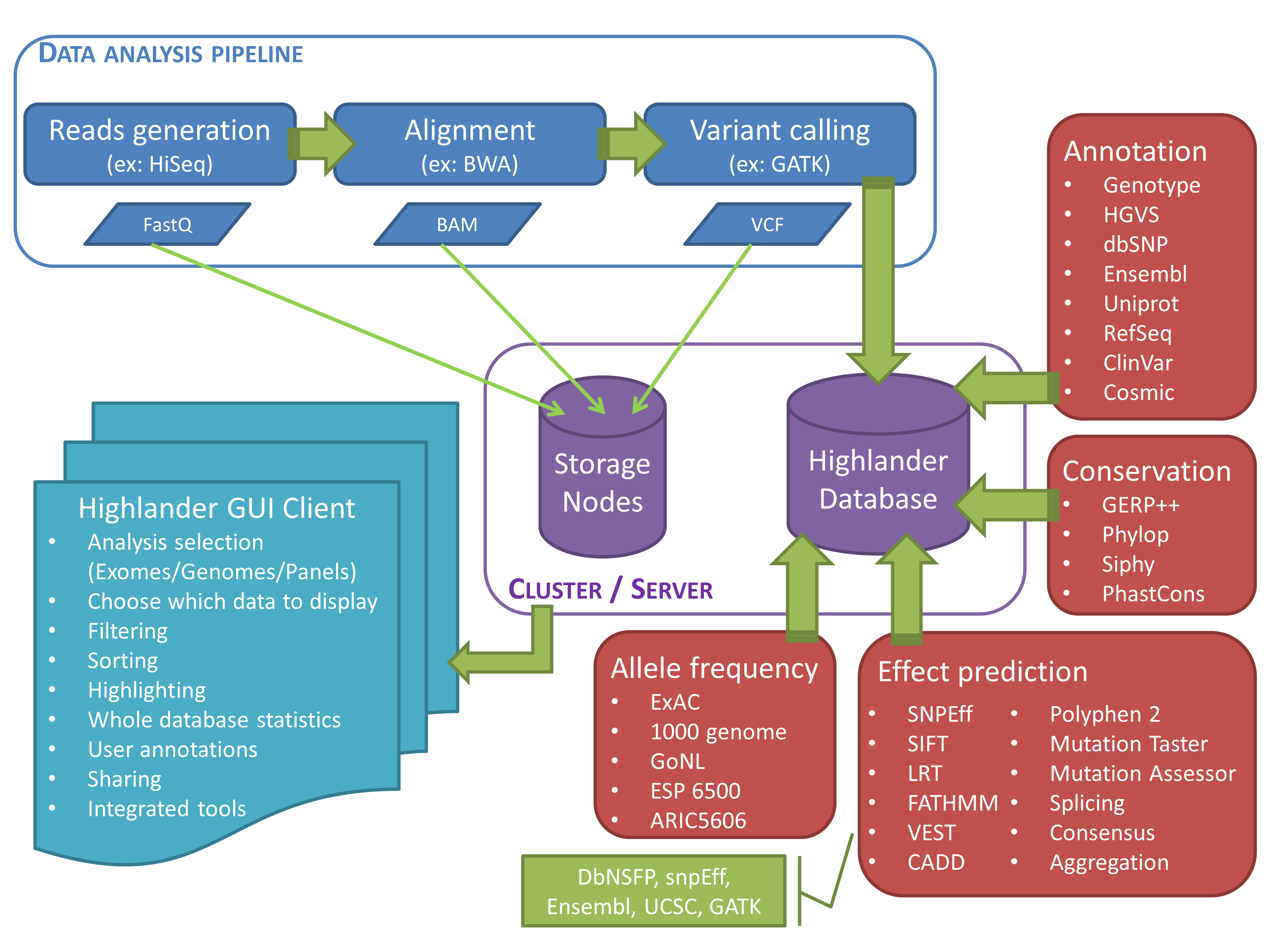
We have developed Highlander, a Java software coupled to a local database, in order to centralize all variant data and annotations from the lab, and to provide powerful filtering tools that are easily accessible to the biologist. Data can be generated by any NGS machine, (such as those of Illumina) and most variant callers (such as Broad Institute's GATK) for SNVs, small INDELs, CNVs (using e.g. ExomeDepth) and other SVs such as gene fusions (using e.g. GRIDSS). Somatic calls are also supported (using e.g. Mutect2). Variant calls are annotated using DBNSFP (providing predictions from 20 different programs, splicing predictions, prioritization scores from CADD or VEST, amongst others), SnpEff (providing topological and functional annotation) and gnomad (providing minor allele frequencies), subsequently imported into the database. The database is used to compute local minor allele frequencies (possibly compartmentalized per pathology), allowing for the discrimination of variants based on their representation in the database. The Highlander GUI easily allows for complex queries to this database, using shortcuts for certain standard criteria, such as "sample-specific variants", "variants common to specific samples" or "combined-heterozygous genes". Users can browse through query results using sorting, masking and highlighting of information. Quality control can be performed with a variety of tools such as FastQC reports, pedigree and sex checker, coverage visualization and histograms displaying various metrics. Highlander also gives access to useful additional tools, including visualization of the alignment, an algorithm that checks all available alignments for allele-calls at specific positions, or access to external tools such as Exomiser or Kraken.
General infrastructure
Highlander multiplatform client
Getting access to Highlander
To use the Highlander client, you need a working local Highlander database (and associated NGS pipeline) which is not provided as a "turnkey solution". Different Highlander databases and pipelines are currently available at UCLouvain (de Duve Institute), Saint-Luc Hospital, Erasme Hospital and UZ Brussel.
If you are interested to join the Highlander consortium, please contact Pr. Miikka Vikkula.
Highlander usage
Highlander is heavily used in our lab and has already lead to published discoveries:
Limaye et al. - Somatic activating PIK3CA mutations cause sporadic venous malformation
The American Journal of Human Genetics 2015, 97:914–921 (doi:10.1016/j.ajhg.2015.11.011)
Soblet et al. - Blue Rubber Bleb Nevus (BRBN) Syndrome is caused by Somatic Double-Mutations in TIE2 Endothelial Receptor Tyrosine Kinase
The Journal of Investigative Dermatology 2017, 137:207-216; (doi:10.1016/j.jid.2016.07.034)
Amyere et al. - Germline Loss-of-Function Mutations in EPHB4 Cause a Second Form of Capillary Malformation-Arteriovenous Malformation (CM-AVM2) Deregulating RAS-MAPK Signaling.
Circulation 2017, 136(11):1037-1048; (doi:10.1161/CIRCULATIONAHA.116.026886)
Brouillard et al. - Loss of ADAMTS3 activity causes lymphangiectasia-lymphedema syndrome 3.
Human Molecular Genetics 2017, ddx297; (doi:10.1093/hmg/ddx297)
Basha et al. - Whole exome sequencing identifies mutations in 10% of patients with familial non-syndromic cleft lip and/or palate in genes mutated in well-known syndromes.
Journal of Medical Genetics 2018, 55:449-458; (doi:10.1136/jmedgenet-2017-105110)
Fastré et al. - Splice-site mutations in VEGFC cause loss-of-function and Nonne-Milroy-like primary lymphedema.
Clinical Genetics 2018; (doi:10.1111/cge.13204)
Demeer et al. - Unmasking familial CPX by WES and identification of novel clinical signs.
American Journal of Medical Genetics Part A 2018, 176A:2661-2667 ; (doi:10.1002/ajmg.a.40630)
Dewulf et al. - SLC13A3 variants cause acute reversible leukoencephalopathy and αKG accumulation.
Annals of Neurology, 2019, 85(3):385-395; (doi:10.1002/ana.25412)
Revencu et al. - RASA1 mosaic mutations in patients with capillary malformation – arteriovenous malformation.
Journal of Medical Genetics, 2019; (doi:10.1136/jmedgenet-2019-106024)
Demeer et al. - Likely Pathogenic Variants in One Third of Non-Syndromic Discontinuous Cleft Lip and Palate Patients.
Genes, 2019, 10(10), 833; (doi:10.3390/genes10100833)
Galot et al. - Liquid biopsy for mutational profiling of locoregional recurrent and/or metastatic head and neck squamous cell carcinoma.
Oral Oncology 2020, 104; (doi:10.1016/j.oraloncology.2020.104631)
Van Marcke et al. - Tumor sequencing is useful to refine the analysis of germline variants in unexplained high-risk breast cancer families.
Breast Cancer Research 2020, 22(36); (doi:10.1186/s13058-020-01273-y)
El-Sibai et al. - Dysregulation of Rho GTPases in orofacial cleft patients-derived primary cells leads to impaired cell migration, a potential cause of cleft/lip palate development.
Cells & Development 2021, 165(203656); (doi:10.1016/j.cdev.2021.203656)
Brouillard et al. - Non‑hotspot PIK3CA mutations are more frequent in CLOVES than in common or combined lymphatic malformations.
Orphanet Journal of Rare Diseases 2021, 16:267; (doi:10.1186/s13023-021-01898-y)
Sepehr et al. - KRAS-driven model of Gorham-Stout disease effectively treated with trametinib.
JCI Insight 2021, 6(15):e149831; (doi:10.1172/jci.insight.149831)
Byrne et al. - Pathogenic variants in MDFIC cause autosomal recessive central conducting lymphatic anomaly.
Science Translational Medicine 2022, 14(634): eabm4869; (doi:10.1126/scitranslmed.abm4869)
Citing Highlander
Highlander (https://sites.uclouvain.be/highlander)