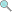3.0 credits
22.5 h + 7.5 h
1q
Teacher(s)
Fustin Charles-André ;
Language
Anglais
Main themes
1. Basic definitions
2. Ionization modes
3. Analyzers
4. Chromatographic couplings
5. Spectral data interpretation
6. Introduction to the identification and sequencing of proteins and peptides by mass spectrometry.
Aims
This course covers technical aspects of mass spectrometry and interpretation of spectral data.
The contribution of this Teaching Unit to the development and command of the skills and learning outcomes of the programme(s) can be accessed at the end of this sheet, in the section entitled “Programmes/courses offering this Teaching Unit”.
Content
After a brief survey of the basic definitions, the various mode of ionization (EI, CI, FAB, ESI, APCI, APPI, DESI, DAPCI, DAPPI, EESI, thermospray, ASAP) will be described in details. The different analyzers (Quad, triple Quad, Trappes, TOF, Orbitrap, FTICR) will be presented together with their possible scanning modes and their combinations. Couplings with Gc and HPLC will be presented. The interpretation of spectral data will first emphasize the differences between low resolution and low accuracy versus high resolution and high accuracy data. The importance of the isotopic cluster will be demonstrated. The principal rules of fragmentation of radical-cations will be presented together with some basic rules for the fragmentation of ions with even-number of electrons. Selected examples and exercises will be explicitly studied in this part of the course. At the end of the course, a short introduction to the use of mass spectrometry for the analysis of proteins and peptides and for the analysis of non-covalente species will be given.
Other information
Prerequisite:
- Basic knowledge of chemistry and physics
- CHM1251C course.
Faculty or entity<