What can R-SWMS do?
R-SWMS is composed of several inter-connected modules, shown on the flow chart below. Each module is explained in the Manual.
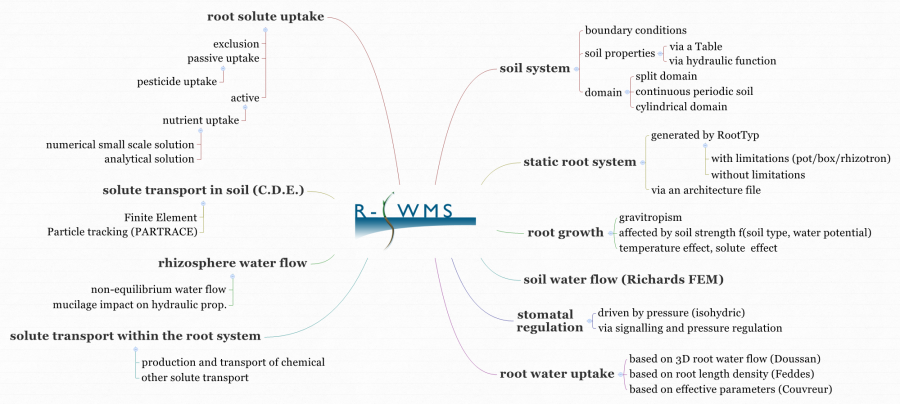
What is needed to install and run R-SWMS?
R-SWMS runs in a linux environment. It has been tested under Ubuntu. The gcc and gfortran free compilers need to be installed. In addition, several free libraries are also needed: openmpi, lapack and mumps.
To vizualise input and output files, you should download the freeware Paraview.
In addition, R-SWMS users have also developped Python or Matlab routines to extract and analyze output files.
R-SWMS examples
Hereafter, you'll find several nice videos showing R-SWMS simulations plotted via paraview.
Example 1:modelling split root experiment with production of ABA
Example 2: Solute transport with root exclusion under day-night transpiration cycles
Example 3: Exclusion, passive and active solute transport modeled with Particle Tracing
Run your own R-SWMS examples
These R-SWMS examples are exercises allowing new users to get used to the input and output files. These examples can also be used to start building your own scenarios. You can find them in the Manual.
- Example 1: Infiltration in a dry soil
- Example 2: A simple root water uptake scenario
- Example 3: A complex root water uptake scenario
- Example 4: Complmex root water uptake with an architecture generated with RootTyp
- Example 5: A growing root taking up water
- Example 6: Hormonal signalling
- Example 7: Solute transport with Partrace